FES-Forward Blog
Explore the Boundaries of Synthetic Biology with us. Stay up-to-date with the latest synthetic biology and DNA synthesis news, updates, and research.
Breaking Down Barriers in DNA Synthesis: High/Low GC Content DNA Sequences
SEP 4, 2024 │ 11 MIN READ
Explore the challenges of synthesizing and assembling high/low GC content DNA sequences including regulatory elements, different bacteria/yeast species, and coding sequences.
GC content, the ratio of guanine (G) and cytosine (C) bases in DNA, plays a crucial role in the stability and functionality of genetic material. However, extremes in GC content—whether high or low—pose significant challenges for DNA synthesis, sequencing, and gene editing technologies. These challenges can hinder the accurate assembly and manipulation of genetic constructs, creating bottlenecks in research and development. In this second installment of our four-part series, we delve into the complexities of GC-rich and GC-poor regions, exploring their natural occurrences and the innovative strategies being developed to overcome the associated obstacles.
What is GC Content?
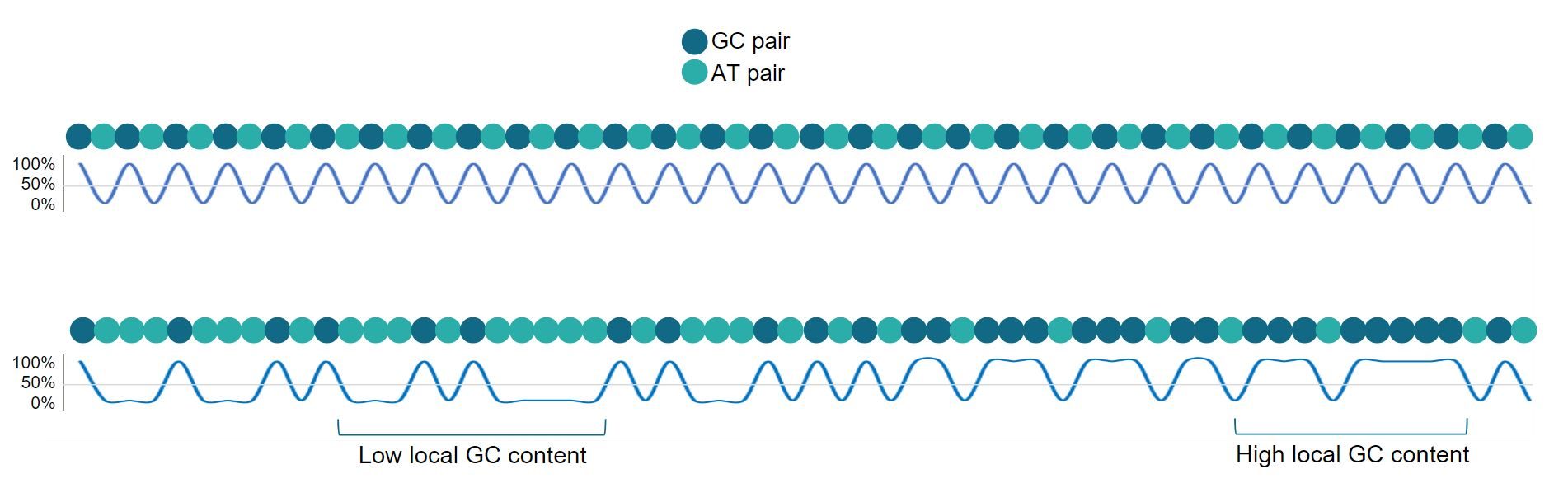
Base composition varies widely between organisms and across genomes, where the proportion of A & T is rarely equal to that of G & C (1). GC content is the percentage of G & C bases within a sequence, where that can be either the global GC content across a while construct or genome, or local GC content within short sequences or specific genes. For example, local GC-rich repeats include ‘CpG islands’, which are often correlated with promoter regions and transcription start sites, while isochores are genomic stretches over 300 kb in length with uniformly high GC and AT content as shown in Figure 1 (2,3).
Why is GC Content Important?
GC content plays a critical role in gene expression, stability, and overall DNA structure. Each of the four bases can bind to their complementary base through hydrogen bonds. Adenosine (A) forms two hydrogen bonds with thymine (T), while guanosine (G) forms three hydrogen bonds with cytosine (C), making the GC pairing stronger due to the extra hydrogen bond (4). This causes them to be more thermally stable and requires higher temperatures for denaturation, making it vital in designing robust genetic constructs. Conversely, low GC content can influence the ease of DNA denaturation and the efficiency of processes like Polymerase Chain Reaction (PCR), impacting research outcomes.
When studying the relationship between gene expression and protein function through genomics, researchers have identified specific patterns in both local and global GC content that correlate with differences in expression, stability, and function. Understanding and optimizing GC content is crucial for applications ranging from synthetic biology to vaccine development. The ability to synthesize DNA with precise GC content allows researchers to fine tune expression and stability of genes underscoring the critical role of GC content in modern biotech applications.
What are Common Examples of Naturally Occurring High or Low GC Content?
High GC
- In vertebrates, important regulatory domains, such as promoters, enhancers, and control elements are frequently GC-rich, leading to their predicted function as sites for transcription factor binding (5). For example, 72% of human promoter sequences are GC rich even though the genome only contains ~ 41% total GC content (6, 7).
- Coding sequences frequently display high GC content including essential housekeeping genes, tumor-suppressor genes, and tissue-specific genes. Several studies have also found significant differences in gene structure between high GC and low GC regions (8).
- CpG islands may play a role in disease development as hot spots of instability, where more repeats can increase the severity of the disease.
- Huntington’s disease is significantly associated with over 30 CpG sites (CAG repeats) in the HTT gene (9).
- Expansion of the (CGG)n motifs in the FMR1 locus from < 62 to > 250 is a known phenotype of Fragile X syndrome (10).
- ALS (Amyotrophic Lateral Sclerosis) being characterized as an expansion of the (GGGGCC)n motif in the C9orf72 gene (11)
Low GC Content
- Some DNA and RNA binding sites have low GC content, including the TATATAA motif that binds the TATA-binding protein (12), and AU-rich elements in the 3’ UTR of transcripts that are targets for RNA-binding proteins (13).
Extreme GC Content
- Bacteria have a much wider range of total genomic GC content with extremes on both ends, such as A. dehalogens (75% overall GC) and C. ruddii (17% overall GC) (14,15,16).
- In yeast, chromatin structures contain characteristic bands of GC rich and AT rich isochores (17).
Why is GC Content Challenging in DNA Synthesis?
There are certain high and low GC content thresholds that can make DNA synthesis, amplification, and sequencing difficult when working with oligonucleotides and gene fragments.
Current DNA synthesis vendors usually consider any sequence with less than 25% GC content to be too low and over 65 – 75% to be too high for accurate synthesis. This is because GC rich regions, with their strong hydrogen bonds and duplex stability, can form secondary structures that are difficult to denature and amplify, hindering accurate synthesis and sequencing. For example, when using current chemical synthesis methods, contiguous d(GC or CG) repeats have a severe effect on performance (18,19).
What are Current Molecular Biology Solutions to Working with High or Low GC Content?
Codon optimization is atechnique some use to normalize the GC content of sequences to fit within the acceptable range for chemical DNA synthesis. Unfortunately, synonymous codon changes may cause unintended consequences at both the mRNA and protein levels (22-24). At the mRNA level, they can result in alternative mRNA folding structures and altered mRNA splicing patterns. In addition, mRNA regulation mechanisms, such as expression control by hybridization of short noncoding microRNAs, can inadvertently be affected by changes to the mRNA sequence. At the protein level, effects can include disruption to pattern of tRNA usage, protein conformation changes, and changes to post translational modifications. Thus, it is desirable to have access to the full repertoire of sequence motifs to best design your studies and to mitigate undesired downstream effects.
Another challenge with working with high or low GC content is amplification through PCR. Polymerase stalling, often triggered by GC rich regions, results in truncated products or skipped regions that lower the purity of the product (20,21). To improve PCR amplification of DNA sequences with high GC content, organic molecules such as dimethyl sulfoxide (DMSO), glycerol, polyethylene glycol, formamide, betaine, 7-deazadGTP, and dUTP can be added to the reaction mixture combined with annealing temperature and cycle time optimization to denature strong secondary structure and prevent polymerase stalling (22). However, these techniques do not work for every sequence and are not solutions for DNA synthesis of GC-rich sequences.
Fully Enzymatic Synthesis (FES) Enables Successful DNA Synthesis of High or Low GC Content
How can long oligos created with Fully Enzymatic Synthesis (FES) help build high and low GC content DNA sequences? Our FES technology was specifically engineered to overcome the challenges associated with synthesizing complex high or low GC sequences, such as those found in CpG islands, GC/AT isochores, and protein binding sites.
Through directed evolution, our proprietary enzymatic synthesis process operates at highly elevated temperatures allowing us to melt secondary structure and synthesize complex DNA. Figure 2 shows the synthesis of the (GGGGCC)30 sequence previously mentioned, thought to be associated with ALS, that consists of a 100% GC 180mer.
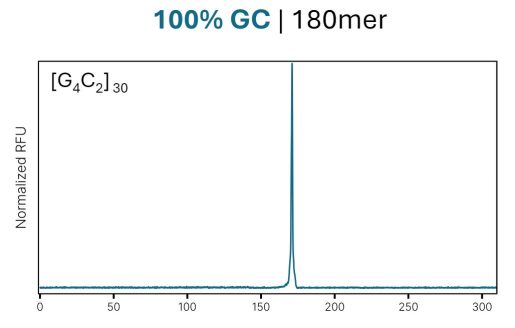
Having the ability to synthesize complex sequences like both high and low GC content, combined with high cycle efficiencies of 99.9% for synthesis of up to 400mer oligonucleotides allows the design flexibility to embed high or low GC content regions in the middle of the oligo to aide in assembly or cloning of these complex sequences. With no limitations on sequence complexity, researchers can reliably order accurate GC-rich and GC-poor sequences with minimal impurities.
FES technology is engineered to overcome these obstacles and provide the precision and reliability needed for success. It has now been proven over a wide variety of complex sequence and molecular cloning/gene assembly techniques.
To learn more about overcoming the challenges of synthesizing high DNA complexity, download our comprehensive white paper or see the first installments of this blog series on Homopolymers and Repetitive Sequences.
Explore More
Download the Complexity White Paper: Explore other complex DNA sequences by downloading our latest white paper on complexity.
Contact Us for More Information: Have questions or need more details? Reach out to our team for expert guidance on your DNA synthesis needs.
References
- Mooers, A. O., & Holmes, E. C. (2000). The evolution of base composition and phylogenetic inference. TREE, 15(9), 365–369. http://www.dbbm.fiocruz.br/james/index.html
- Bernardi, G. (1993). The Vertebrate Genome: Isochores and Evolution. Molecular Biology and Evolution, 10(1), 186–204. https://academic.oup.com/mbe/article/10/1/186/1030039
- Illingworth, R. S., & Bird, A. P. (2009). CpG islands – “A rough guide.” In FEBS Letters (Vol. 583, Issue 11, pp. 1713–1720). https://doi.org/10.1016/j.febslet.2009.04.012
- Harding, S. E., Channell, G., & Phillips-Jones, M. K. (2018). The discovery of hydrogen bonds in DNA and a re-evaluation of the 1948 Creeth two-chain model for its structure. In Biochemical Society Transactions (Vol. 46, Issue 5, pp. 1171–1182). Portland Press Ltd. https://doi.org/10.1042/BST20180158
- Jaksik, R., & Rzeszowska-Wolny, J. (2012). The distribution of GC nucleotides and regulatory sequence motifs in genes and their adjacent sequences. Gene, 492(2), 375–381. https://doi.org/10.1016/j.gene.2011.10.050
- Saxonov, Serge, Paul Berg, and Douglas L. Brutlag. “A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters.” Proceedings of the National Academy of Sciences 103.5 (2006): 1412-1417.
- Piovesan, A., Pelleri, M. C., Antonaros, F., Strippoli, P., Caracausi, M., & Vitale, L. (2019). On the length, weight and GC content of the human genome. BMC Research Notes, 12(1). https://doi.org/10.1186/s13104-019-4137-z
- Amit, M., Donyo, M., Hollander, D., Goren, A., Kim, E., Gelfman, S., Lev-Maor, G., Burstein, D., Schwartz, S., Postolsky, B., Pupko, T., & Ast, G. (2012). Differential GC Content between Exons and Introns Establishes Distinct Strategies of Splice-Site Recognition. Cell Reports, 1(5), 543–556. https://doi.org/10.1016/j.celrep.2012.03.013
- Lu, A. T., Narayan, P., Grant, M. J., Langfelder, P., Wang, N., Kwak, S., Wilkinson, H., Chen, R. Z., Chen, J., Simon Bawden, C., Rudiger, S. R., Ciosi, M., Chatzi, A., Maxwell, A., Hore, T. A., Aaronson, J., Rosinski, J., Preiss, A., Vogt, T. F., … Horvath, S. (2020). DNA methylation study of Huntington’s disease and motor progression in patients and in animal models. Nature Communications, 11(1). https://doi.org/10.1038/s41467-020-18255-5
- Casas-Delucchi, C. S., Daza-Martin, M., Williams, S. L., & Coster, G. (2022). The mechanism of replication stalling and recovery within repetitive DNA. Nature Communications, 13(1). https://doi.org/10.1038/s41467-022-31657-x
- Depienne, C., & Mandel, J. L. (2021). 30 years of repeat expansion disorders: What have we learned and what are the remaining challenges?. The American Journal of Human Genetics, 108(5), 764-785.
- Wong, J. M., & Bateman, E. (1994). TBP-DNA interactions in the minor groove discriminate between A:T and T:A base pairs. In Nucleic Acids Research (Vol. 22, Issue 10). https://academic.oup.com/nar/article/22/10/1890/1248693
- Barreau, C., Paillard, L., & Osborne, H. B. (2005). AU-rich elements and associated factors: Are there unifying principles? In Nucleic Acids Research (Vol. 33, Issue 22, pp. 7138–7150). https://doi.org/10.1093/nar/gki1012
- Hildebrand, F., Meyer, A., & Eyre-Walker, A. (2010). Evidence of selection upon genomic GC-content in bacteria. PLoS genetics, 6(9), e1001107
- Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006 Oct 13;314(5797):267. doi: 10.1126/science.1134196. PMID: 17038615.
- https://microbewiki.kenyon.edu/index.php/Anaeromyxobacter_dehalogenans
- Dekker, J. GC- and AT-rich chromatin domains differ in conformation and histone modification status and are differentially modulated by Rpd3p. Genome Biol 8, R116 (2007). https://doi.org/10.1186/gb-2007-8-6-r116
- Li, Q., & Yan, H. (2023). “Difficult” deoxyribonucleotide sequences in the solid-phase synthesis by the phosphoramidite chemistry. Nucleosides, Nucleotides & Nucleic Acids, 43(7), 655–663. https://doi.org/10.1080/15257770.2023.2295478
- Hachmann, J. P., & Lebl, M. (2006). Synthesis of Poly d(G-C) Oligonucleotides. Nucleosides, Nucleotides & Nucleic Acids, 25(7), 705–717. https://doi.org/10.1080/15257770600725903
- Green MR, Sambrook J. Polymerase Chain Reaction (PCR) Amplification of GC-Rich Templates. Cold Spring Harb Protoc. 2019 Feb 1;2019(2). doi: 10.1101/pdb.prot095141. PMID: 30710022.
- Voineagu, I., Narayanan, V., Lobachev, K. S., & Mirkin, S. M. (2008). Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proceedings of the National Academy of Sciences, 105(29), 9936-9941.
- Mamedov, T. G., Pienaar, E., Whitney, S. E., TerMaat, J. R., Carvill, G., Goliath, R., Subramanian, A., & Viljoen, H. J. (2008). A fundamental study of the PCR amplification of GC-rich DNA templates. Computational Biology and Chemistry, 32(6), 452–457. https://doi.org/10.1016/j.compbiolchem.2008.07.021
- Lin, B.C., Katneni, U., Jankowska, K.I. et al. In silico methods for predicting functional synonymous variants. Genome Biol 24, 126 (2023). https://doi.org/10.1186/s13059-023-02966-1
- Mauro, V.P. Codon Optimization in the Production of Recombinant Biotherapeutics: Potential Risks and Considerations. BioDrugs 32, 69–81 (2018). https://doi.org/10.1007/s40259-018-0261-x
- Mauro, V. P., & Chappell, S. A. (2014). A critical analysis of codon optimization in human therapeutics. Trends in molecular medicine, 20(11), 604-613.
